Quickstart to ML with dabl¶
Let’s dive right in!
Let’s start with the classic. You have the titanic.csv file and want to predict whether a passenger survived or not based on the information about the passenger in that file. We know, for tabular data like this, pandas is our friend. Clearly we need to start with loading our data:
>>> import pandas as pd
>>> import dabl
>>> titanic = pd.read_csv(dabl.datasets.data_path("titanic.csv"))
Let’s familiarize ourself with the data a bit; what’s the shape, what are the columns, what do they look like?
>>> titanic.shape
(1309, 14)
>>> titanic.head()
pclass survived ... body home.dest
0 1 1 ... ? St Louis, MO
1 1 1 ... ? Montreal, PQ / Chesterville, ON
2 1 0 ... ? Montreal, PQ / Chesterville, ON
3 1 0 ... 135 Montreal, PQ / Chesterville, ON
4 1 0 ... ? Montreal, PQ / Chesterville, ON
[5 rows x 14 columns]
So far so good! There’s already a bunch of things going on in the data that we can see here, but let’s ask dabl what it thinks by cleaning up the data:
>>> titanic_clean = dabl.clean(titanic, verbose=0)
This provides us with lots of information about what is happening in the different columns. In this case, we might have been able to figure this out quickly from the call to head, but in larger datasets this might be a bit tricky. For example we can see that there are several dirty columns with “?” in it. This is probably a marker for a missing value and we could go back and fix our parsing of the CSV, but let’s try and continue with what dabl is doing automatically for now. In dabl, we can also get a best guess of the column types in a convenient format:
>>> types = dabl.detect_types(titanic_clean)
>>> print(types)
continuous dirty_float ... free_string useless
pclass False False ... False False
survived False False ... False False
name False False ... True False
sex False False ... False False
sibsp False False ... False False
parch False False ... False False
ticket False False ... True False
cabin False False ... True False
embarked False False ... False False
boat False False ... False False
home.dest False False ... True False
age_? False False ... False False
age_dabl_continuous True False ... False False
fare_? False False ... False True
fare_dabl_continuous True False ... False False
body_? False False ... False False
body_dabl_continuous True False ... False False
[17 rows x 8 columns]
Having a very rough idea of the shape of our data, we can now start looking at the actual content. The easiest way to do that is using visualization of univariate and bivariate patterns. With plot, we can create plot of the features deemed most important for our task.
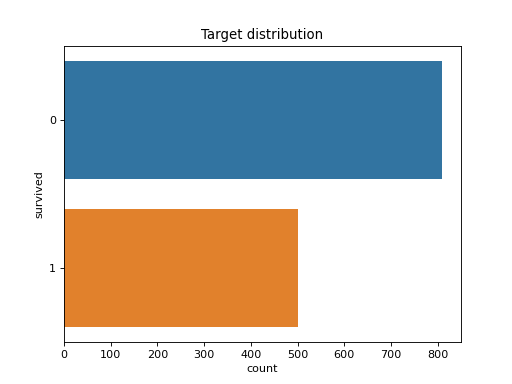
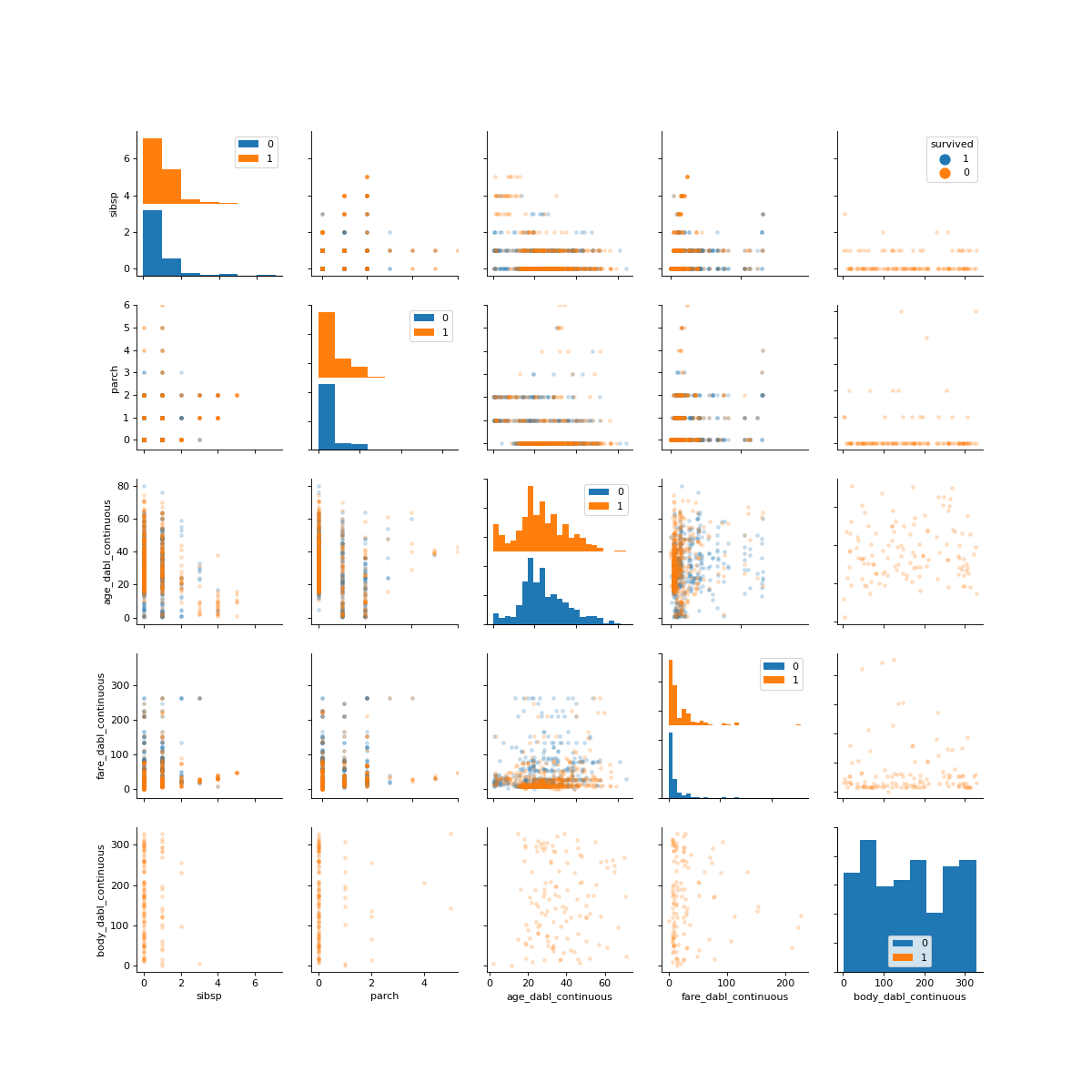
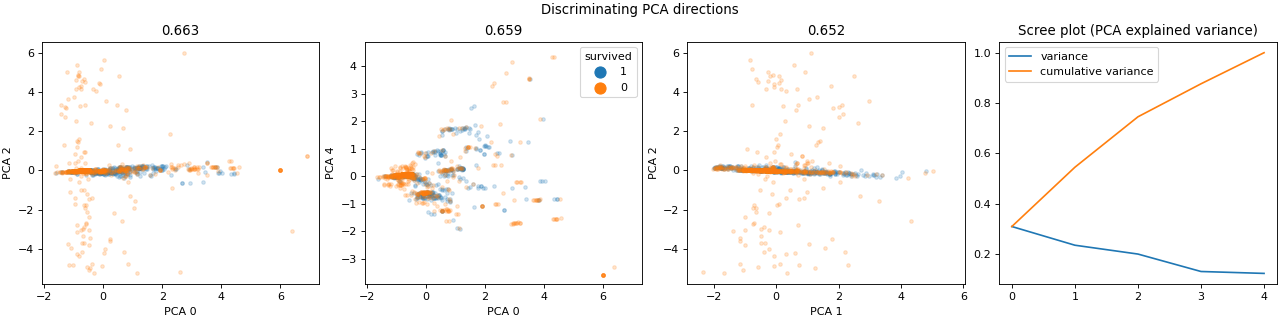
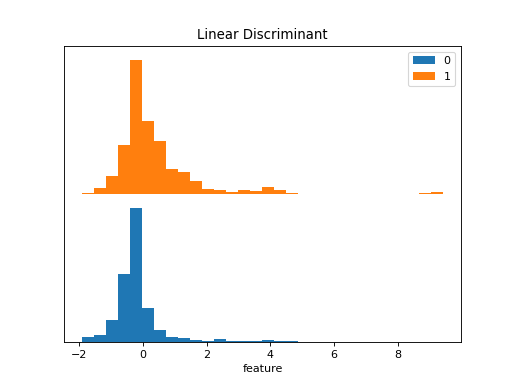
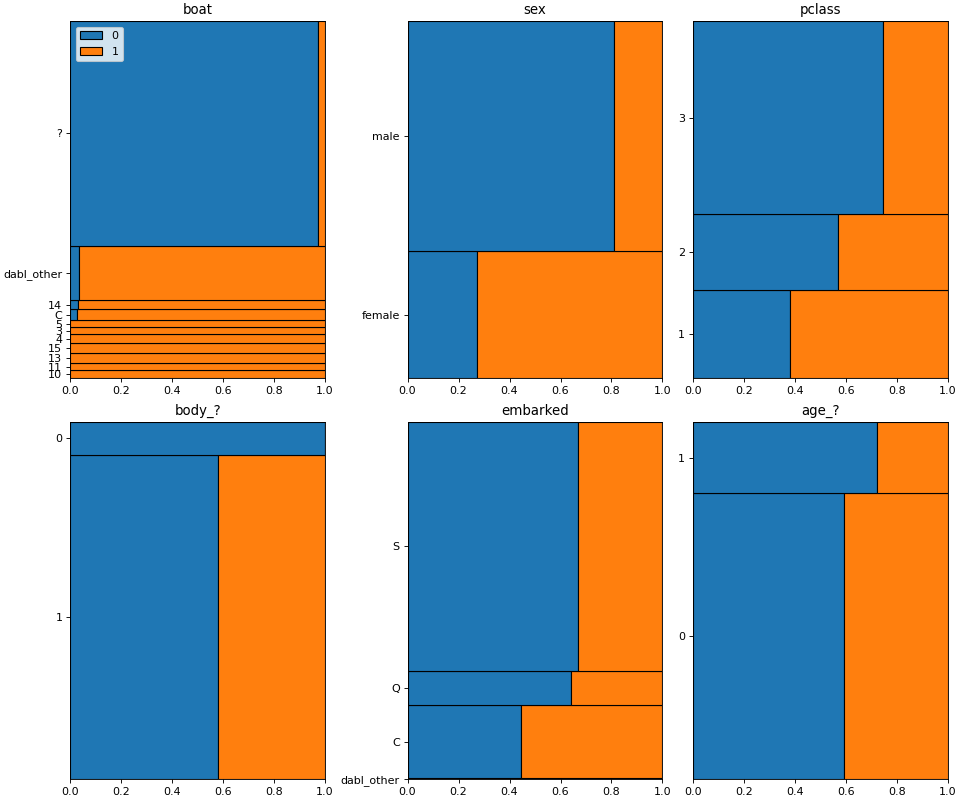
>>> dabl.plot(titanic, 'survived')
Target looks like classification
Linear Discriminant Analysis training set score: 0.578
Finally, we can find an initial model for our data. The SimpleClassifier does all the work for us. It implements the familiar scikit-learn API of fit and predict. Alternatively we could also use the same interface as before and pass the whole data frame and specify the target column.
>>> fc = dabl.SimpleClassifier(random_state=0)
>>> X = titanic_clean.drop("survived", axis=1)
>>> y = titanic_clean.survived
>>> fc.fit(X, y)
DummyClassifier(strategy='prior')
accuracy: 0.618 average_precision: 0.382 recall_macro: 0.500 roc_auc: 0.500
new best (using recall_macro):
accuracy 0.618
average_precision 0.382
recall_macro 0.500
roc_auc 0.500
Name: DummyClassifier(strategy='prior'), dtype: float64
GaussianNB()
accuracy: 0.897 average_precision: 0.870 recall_macro: 0.902 roc_auc: 0.919
new best (using recall_macro):
accuracy 0.897
average_precision 0.870
recall_macro 0.902
roc_auc 0.919
Name: GaussianNB(), dtype: float64
MultinomialNB()
accuracy: 0.888 average_precision: 0.981 recall_macro: 0.891 roc_auc: 0.985
DecisionTreeClassifier(class_weight='balanced', max_depth=1)
accuracy: 0.976 average_precision: 0.954 recall_macro: 0.971 roc_auc: 0.971
new best (using recall_macro):
accuracy 0.976
average_precision 0.954
recall_macro 0.971
roc_auc 0.971
Name: DecisionTreeClassifier(class_weight='balanced', max_depth=1), dtype: float64
DecisionTreeClassifier(class_weight='balanced', max_depth=5)
accuracy: 0.957 average_precision: 0.943 recall_macro: 0.953 roc_auc: 0.970
DecisionTreeClassifier(class_weight='balanced', min_impurity_decrease=0.01)
accuracy: 0.976 average_precision: 0.954 recall_macro: 0.971 roc_auc: 0.971
LogisticRegression(C=0.1, class_weight='balanced')
accuracy: 0.963 average_precision: 0.986 recall_macro: 0.961 roc_auc: 0.989
Best model:
DecisionTreeClassifier(class_weight='balanced', max_depth=1)
Best Scores:
accuracy 0.976
average_precision 0.954
recall_macro 0.971
roc_auc 0.971
Name: DecisionTreeClassifier(class_weight='balanced', max_depth=1), dtype: float64
SimpleClassifier(random_state=0, refit=True, verbose=1)